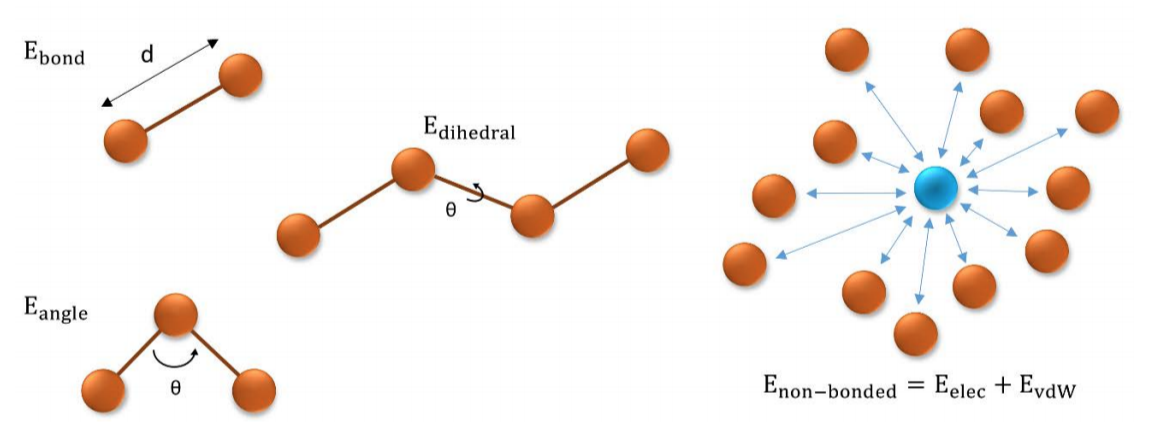
Small molecule force field parametrization for atomistic Molecular Dynamics simulations — HBP Brain Simulation Platform Guidebook 0.1.1 documentation

Fixed-Charge Atomistic Force Fields for Molecular Dynamics Simulations in the Condensed Phase: An Overview | Journal of Chemical Information and Modeling

John Chodera (he/him) on Twitter: "Typical molecular mechanics (MM) force fields represent the potential energy function of a biomolecular system with extremely simple, decoupled algebraic (mostly harmonic) expressions that can't deal with

All-Atom Force Field for Molecular Dynamics Simulations on Organotransition Metal Solids and Liquids. Application to M(CO)n (M = Cr, Fe, Ni, Mo, Ru, or W) Compounds | The Journal of Physical Chemistry
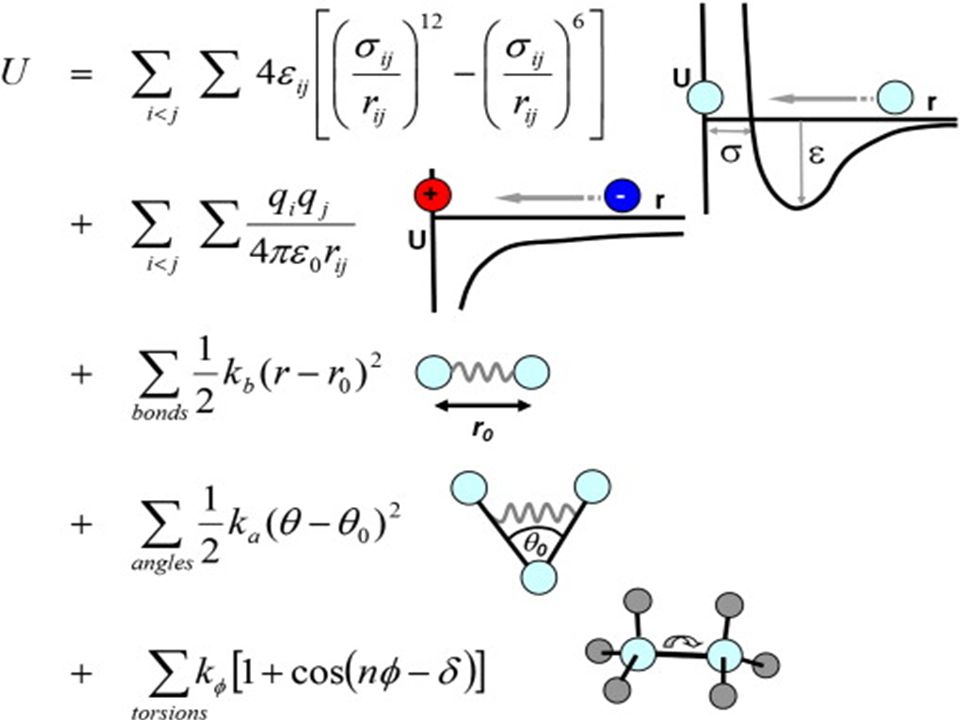
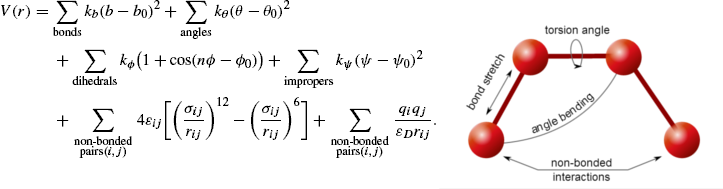
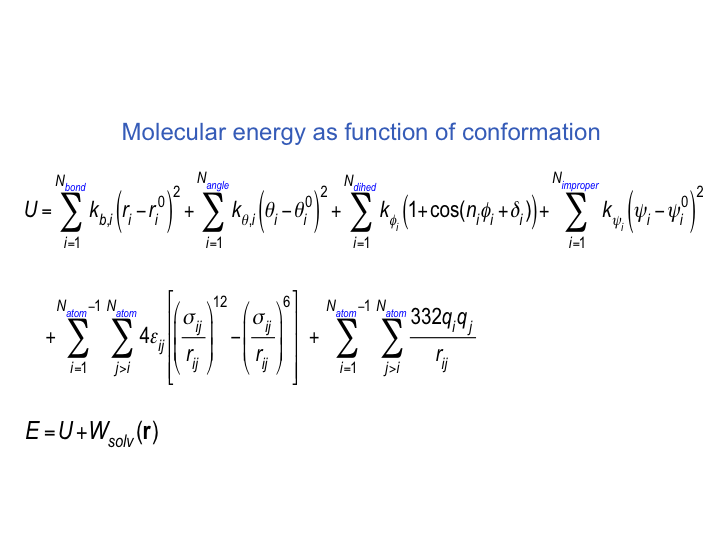
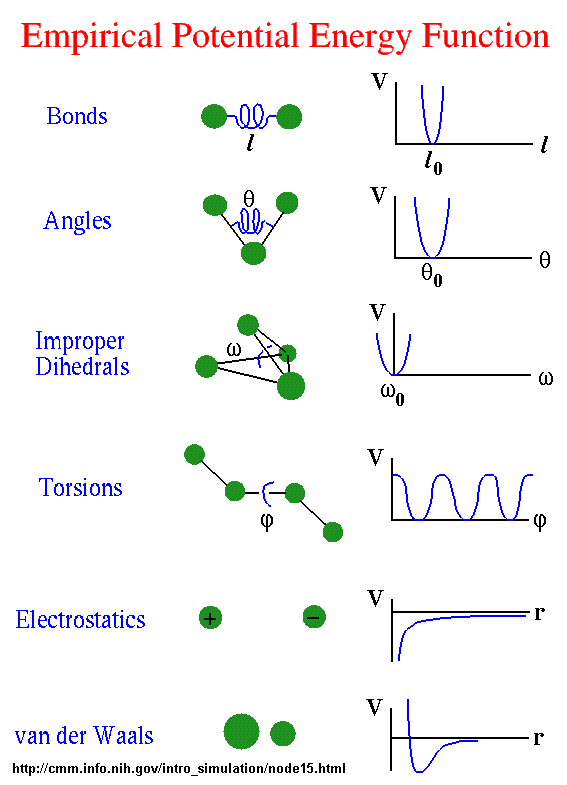
Molecular Mechanics a. Force fields b. Energy minimization / Geometry optimization c. Molecular mechanics examples. - ppt download

PPT – Molecular Mechanics Force Field Method PowerPoint presentation | free to view - id: 1158d2-NDA5Z